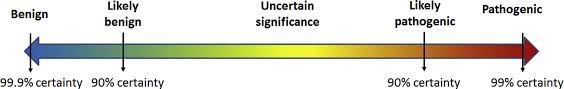
Para poder establecer la causalidad de una variante genética respecto de un fenotipo, es necesario obtener evidencia científica suficiente. La clasificación de patogenicidad propuesta por la ACMG establece 5 categorías de clasificación: Benigna, probablemente benigna, variante de significado incierto, probablemente patogénica y patogénica. Estas categorias están definidas en función de la probabilidad de que exista una relación de causalidad entre la variante detectada y el fenotipo, de forma que una variante clasificada como probablemente patogénica tiene un 90% de certeza de ser patogénica, y una clasificada como patogénica tiene un 99% de probabilidad de serlo realmente. Esto también significa, que clasificar una variante como VSI significa que la probabilidad de que la variante sea patogénica se sitúa en algún punto entre el 11 y el 89% de probabilidad.
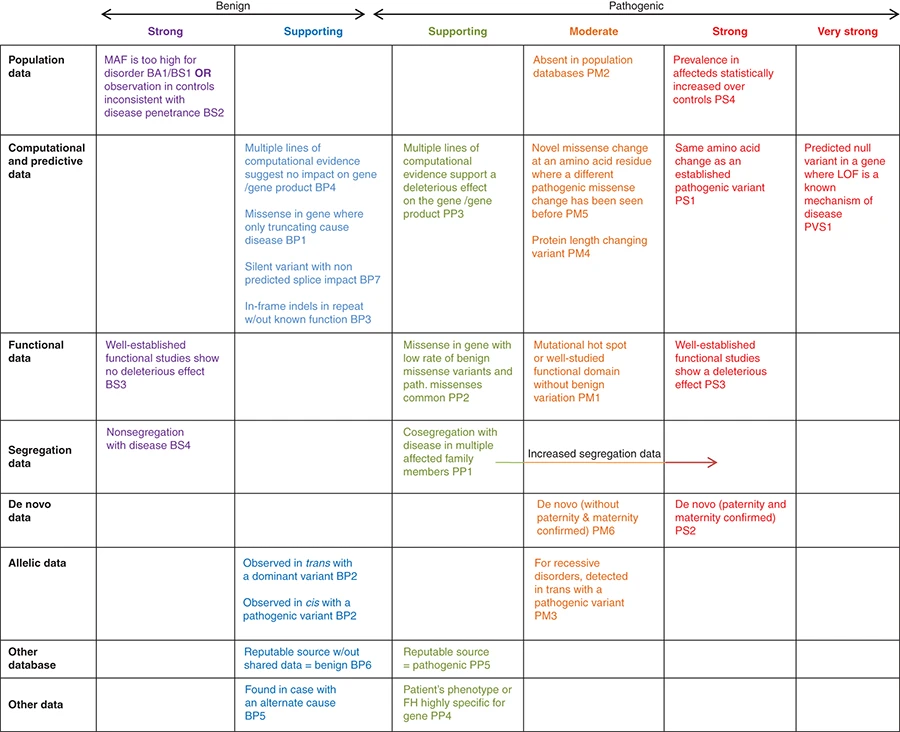
También ha propuesto un sistema de análisis de los distintos tipos de evidencia, desde los datos obtenidos en estudios poblacionales, los datos de análisis funcional, predicción in silico o estudio de segregación, y los ha clasificado en función del peso relativo de cada tipo de datos en la atribución de causalidad (evidencia muy fuerte, fuerte, moderada, de apoyo (supporting) o no suficiente por si misma).
Existen varias bases de datos en las que se puede consultar la patogenicidad de las variantes identificadas en un estudio de secuenciación, si es que han sido descritas en la bibliografía con anterioridad y se han realizado investigaciones sobre la causalidad. ClinGen es la base de datos aprobada por la FDA para este propósito.
En el caso de que las variantes no hayan sido investigadas con anterioridad, será necesario revisar las bases de datos poblacionales, tanto de personas sanas como de variantes patogénicas, así como otros tipos de evidencia. Varsome es una herramienta web que utiliza múltiples bases de datos (incluida ClinGen) para proporcionar una interpretación automatizada siguiendo los criterios del ACMG.
332710
{332710:8R2WCUBM},{332710:N25CS3PK}
vancouver
50
6113
https://neuropediatoolkit.org/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3A%22zotpress-84260f9b06724dd227f249e328fa2ca3%22%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22N25CS3PK%22%2C%22library%22%3A%7B%22id%22%3A332710%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Richards%20et%20al.%22%2C%22parsedDate%22%3A%222015-05%22%2C%22numChildren%22%3A6%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%201.35%3B%20%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%20style%3D%5C%22clear%3A%20left%3B%20%5C%22%3E%5Cn%20%20%20%20%3Cdiv%20class%3D%5C%22csl-left-margin%5C%22%20style%3D%5C%22float%3A%20left%3B%20padding-right%3A%200.5em%3B%20text-align%3A%20right%3B%20width%3A%201em%3B%5C%22%3E1.%3C%5C%2Fdiv%3E%3Cdiv%20class%3D%5C%22csl-right-inline%5C%22%20style%3D%5C%22margin%3A%200%20.4em%200%201.5em%3B%5C%22%3ERichards%20S%2C%20Aziz%20N%2C%20Bale%20S%2C%20Bick%20D%2C%20Das%20S%2C%20Gastier-Foster%20J%2C%20et%20al.%20Standards%20and%20Guidelines%20for%20the%20Interpretation%20of%20Sequence%20Variants%3A%20A%20Joint%20Consensus%20Recommendation%20of%20the%20American%20College%20of%20Medical%20Genetics%20and%20Genomics%20and%20the%20Association%20for%20Molecular%20Pathology.%20Genet%20Med%20%5BInternet%5D.%202015%20May%20%5Bcited%202016%20Sep%206%5D%3B17%285%29%3A405%26%23x2013%3B24.%20Available%20from%3A%20%3Ca%20href%3D%27http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpmc%5C%2Farticles%5C%2FPMC4544753%5C%2F%27%3Ehttp%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpmc%5C%2Farticles%5C%2FPMC4544753%5C%2F%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%20%20%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Standards%20and%20Guidelines%20for%20the%20Interpretation%20of%20Sequence%20Variants%3A%20A%20Joint%20Consensus%20Recommendation%20of%20the%20American%20College%20of%20Medical%20Genetics%20and%20Genomics%20and%20the%20Association%20for%20Molecular%20Pathology%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sue%22%2C%22lastName%22%3A%22Richards%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nazneen%22%2C%22lastName%22%3A%22Aziz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sherri%22%2C%22lastName%22%3A%22Bale%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22David%22%2C%22lastName%22%3A%22Bick%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Soma%22%2C%22lastName%22%3A%22Das%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julie%22%2C%22lastName%22%3A%22Gastier-Foster%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Wayne%20W.%22%2C%22lastName%22%3A%22Grody%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Madhuri%22%2C%22lastName%22%3A%22Hegde%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elaine%22%2C%22lastName%22%3A%22Lyon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elaine%22%2C%22lastName%22%3A%22Spector%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karl%22%2C%22lastName%22%3A%22Voelkerding%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Heidi%20L.%22%2C%22lastName%22%3A%22Rehm%22%7D%5D%2C%22abstractNote%22%3A%22The%20American%20College%20of%20Medical%20Genetics%20and%20Genomics%20%28ACMG%29%20previously%20developed%20guidance%20for%20the%20interpretation%20of%20sequence%20variants.%20In%20the%20past%20decade%2C%20sequencing%20technology%20has%20evolved%20rapidly%20with%20the%20advent%20of%20high-throughput%20next%20generation%20sequencing.%20By%20adopting%20and%20leveraging%20next%20generation%20sequencing%2C%20clinical%20laboratories%20are%20now%20performing%20an%20ever%20increasing%20catalogue%20of%20genetic%20testing%20spanning%20genotyping%2C%20single%20genes%2C%20gene%20panels%2C%20exomes%2C%20genomes%2C%20transcriptomes%20and%20epigenetic%20assays%20for%20genetic%20disorders.%20By%20virtue%20of%20increased%20complexity%2C%20this%20paradigm%20shift%20in%20genetic%20testing%20has%20been%20accompanied%20by%20new%20challenges%20in%20sequence%20interpretation.%20In%20this%20context%2C%20the%20ACMG%20convened%20a%20workgroup%20in%202013%20comprised%20of%20representatives%20from%20the%20ACMG%2C%20the%20Association%20for%20Molecular%20Pathology%20%28AMP%29%20and%20the%20College%20of%20American%20Pathologists%20%28CAP%29%20to%20revisit%20and%20revise%20the%20standards%20and%20guidelines%20for%20the%20interpretation%20of%20sequence%20variants.%20The%20group%20consisted%20of%20clinical%20laboratory%20directors%20and%20clinicians.%20This%20report%20represents%20expert%20opinion%20of%20the%20workgroup%20with%20input%20from%20ACMG%2C%20AMP%20and%20CAP%20stakeholders.%20These%20recommendations%20primarily%20apply%20to%20the%20breadth%20of%20genetic%20tests%20used%20in%20clinical%20laboratories%20including%20genotyping%2C%20single%20genes%2C%20panels%2C%20exomes%20and%20genomes.%20This%20report%20recommends%20the%20use%20of%20specific%20standard%20terminology%3A%20%5Cu2018pathogenic%5Cu2019%2C%20%5Cu2018likely%20pathogenic%5Cu2019%2C%20%5Cu2018uncertain%20significance%5Cu2019%2C%20%5Cu2018likely%20benign%5Cu2019%2C%20and%20%5Cu2018benign%5Cu2019%20to%20describe%20variants%20identified%20in%20Mendelian%20disorders.%20Moreover%2C%20this%20recommendation%20describes%20a%20process%20for%20classification%20of%20variants%20into%20these%20five%20categories%20based%20on%20criteria%20using%20typical%20types%20of%20variant%20evidence%20%28e.g.%20population%20data%2C%20computational%20data%2C%20functional%20data%2C%20segregation%20data%2C%20etc.%29.%20Because%20of%20the%20increased%20complexity%20of%20analysis%20and%20interpretation%20of%20clinical%20genetic%20testing%20described%20in%20this%20report%2C%20the%20ACMG%20strongly%20recommends%20that%20clinical%20molecular%20genetic%20testing%20should%20be%20performed%20in%20a%20CLIA-approved%20laboratory%20with%20results%20interpreted%20by%20a%20board-certified%20clinical%20molecular%20geneticist%20or%20molecular%20genetic%20pathologist%20or%20equivalent.%22%2C%22date%22%3A%222015-5%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fgim.2015.30%22%2C%22ISSN%22%3A%221098-3600%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpmc%5C%2Farticles%5C%2FPMC4544753%5C%2F%22%2C%22collections%22%3A%5B%227BFFA6NH%22%2C%22FCKFQGK8%22%2C%22LZWEG8HJ%22%5D%2C%22dateModified%22%3A%222019-07-23T17%3A59%3A35Z%22%7D%7D%2C%7B%22key%22%3A%228R2WCUBM%22%2C%22library%22%3A%7B%22id%22%3A332710%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22MacArthur%20et%20al.%22%2C%22parsedDate%22%3A%222014%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%201.35%3B%20%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%20style%3D%5C%22clear%3A%20left%3B%20%5C%22%3E%5Cn%20%20%20%20%3Cdiv%20class%3D%5C%22csl-left-margin%5C%22%20style%3D%5C%22float%3A%20left%3B%20padding-right%3A%200.5em%3B%20text-align%3A%20right%3B%20width%3A%201em%3B%5C%22%3E1.%3C%5C%2Fdiv%3E%3Cdiv%20class%3D%5C%22csl-right-inline%5C%22%20style%3D%5C%22margin%3A%200%20.4em%200%201.5em%3B%5C%22%3EMacArthur%20DG%2C%20Manolio%20TA%2C%20Dimmock%20DP%2C%20Rehm%20HL%2C%20Shendure%20J%2C%20Abecasis%20GR%2C%20et%20al.%20Guidelines%20for%20investigating%20causality%20of%20sequence%20variants%20in%20human%20disease.%20Nature%20%5BInternet%5D.%202014%20Abril%20%5Bcited%202015%20Jun%2023%5D%3B508%287497%29%3A469%26%23x2013%3B76.%20Available%20from%3A%20%3Ca%20href%3D%27http%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fnature%5C%2Fjournal%5C%2Fv508%5C%2Fn7497%5C%2Ffull%5C%2Fnature13127.html%27%3Ehttp%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fnature%5C%2Fjournal%5C%2Fv508%5C%2Fn7497%5C%2Ffull%5C%2Fnature13127.html%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%20%20%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Guidelines%20for%20investigating%20causality%20of%20sequence%20variants%20in%20human%20disease%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20G.%22%2C%22lastName%22%3A%22MacArthur%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22T.%20A.%22%2C%22lastName%22%3A%22Manolio%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20P.%22%2C%22lastName%22%3A%22Dimmock%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22H.%20L.%22%2C%22lastName%22%3A%22Rehm%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.%22%2C%22lastName%22%3A%22Shendure%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%20R.%22%2C%22lastName%22%3A%22Abecasis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20R.%22%2C%22lastName%22%3A%22Adams%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R.%20B.%22%2C%22lastName%22%3A%22Altman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20E.%22%2C%22lastName%22%3A%22Antonarakis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22E.%20A.%22%2C%22lastName%22%3A%22Ashley%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.%20C.%22%2C%22lastName%22%3A%22Barrett%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20G.%22%2C%22lastName%22%3A%22Biesecker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20F.%22%2C%22lastName%22%3A%22Conrad%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%20M.%22%2C%22lastName%22%3A%22Cooper%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%20J.%22%2C%22lastName%22%3A%22Cox%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%20J.%22%2C%22lastName%22%3A%22Daly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%20B.%22%2C%22lastName%22%3A%22Gerstein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%20B.%22%2C%22lastName%22%3A%22Goldstein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.%20N.%22%2C%22lastName%22%3A%22Hirschhorn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20M.%22%2C%22lastName%22%3A%22Leal%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%20A.%22%2C%22lastName%22%3A%22Pennacchio%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J.%20A.%22%2C%22lastName%22%3A%22Stamatoyannopoulos%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20R.%22%2C%22lastName%22%3A%22Sunyaev%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D.%22%2C%22lastName%22%3A%22Valle%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22B.%20F.%22%2C%22lastName%22%3A%22Voight%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22W.%22%2C%22lastName%22%3A%22Winckler%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C.%22%2C%22lastName%22%3A%22Gunter%22%7D%5D%2C%22abstractNote%22%3A%22The%20discovery%20of%20rare%20genetic%20variants%20is%20accelerating%2C%20and%20clear%20guidelines%20for%20distinguishing%20disease-causing%20sequence%20variants%20from%20the%20many%20potentially%20functional%20variants%20present%20in%20any%20human%20genome%20are%20urgently%20needed.%20Without%20rigorous%20standards%20we%20risk%20an%20acceleration%20of%20false-positive%20reports%20of%20causality%2C%20which%20would%20impede%20the%20translation%20of%20genomic%20research%20findings%20into%20the%20clinical%20diagnostic%20setting%20and%20hinder%20biological%20understanding%20of%20disease.%20Here%20we%20discuss%20the%20key%20challenges%20of%20assessing%20sequence%20variants%20in%20human%20disease%2C%20integrating%20both%20gene-level%20and%20variant-level%20support%20for%20causality.%20We%20propose%20guidelines%20for%20summarizing%20confidence%20in%20variant%20pathogenicity%20and%20highlight%20several%20areas%20that%20require%20further%20resource%20development.%22%2C%22date%22%3A%22Abril%2024%2C%202014%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1038%5C%2Fnature13127%22%2C%22ISSN%22%3A%220028-0836%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.nature.com%5C%2Fnature%5C%2Fjournal%5C%2Fv508%5C%2Fn7497%5C%2Ffull%5C%2Fnature13127.html%22%2C%22collections%22%3A%5B%227BFFA6NH%22%5D%2C%22dateModified%22%3A%222015-07-30T22%3A22%3A13Z%22%7D%7D%5D%7D
1.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med [Internet]. 2015 May [cited 2016 Sep 6];17(5):405–24. Available from:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4544753/