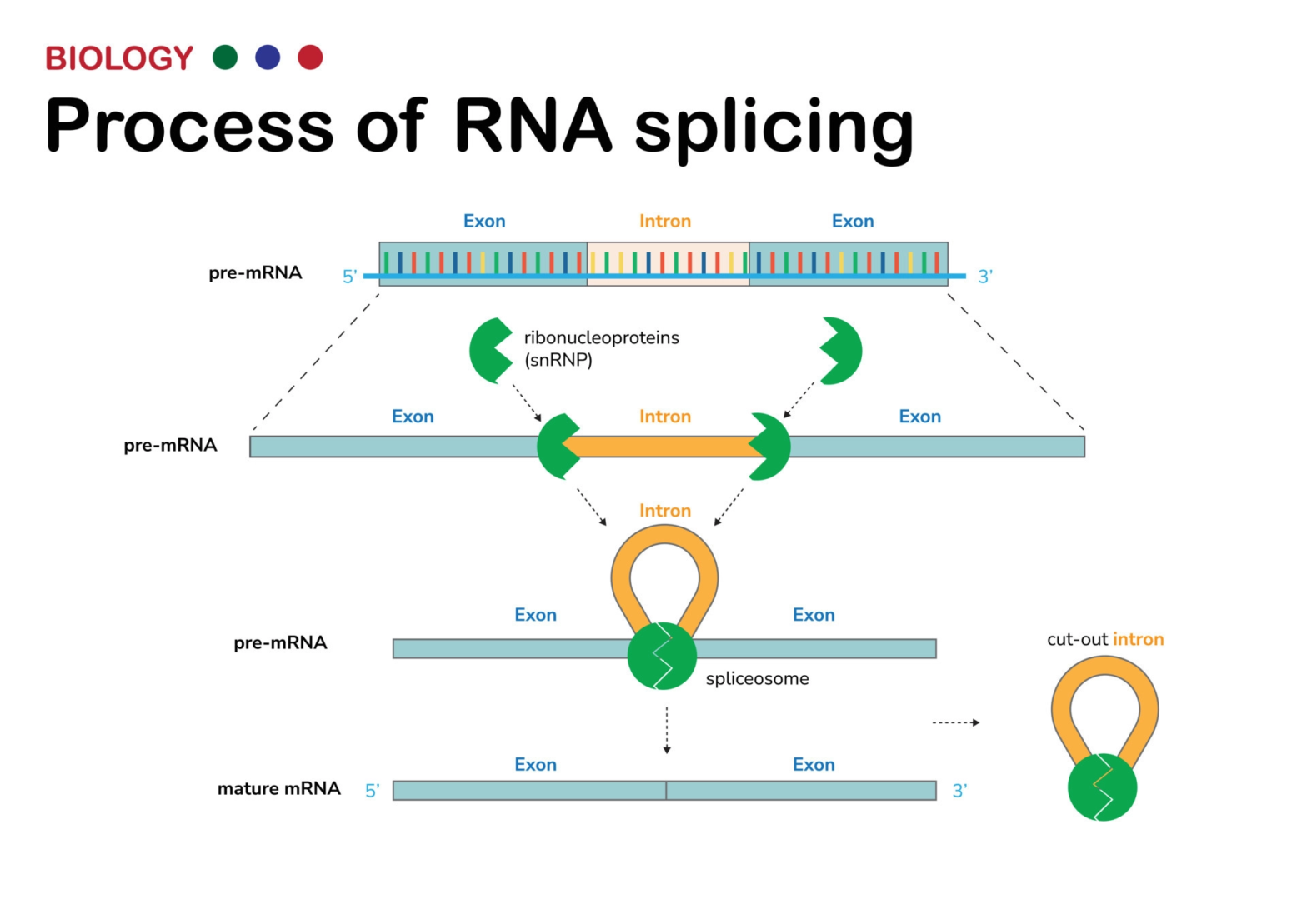
Existen múltiples enfermedades que directa o indirectamente afectan al splicing, por varios mecanismos.
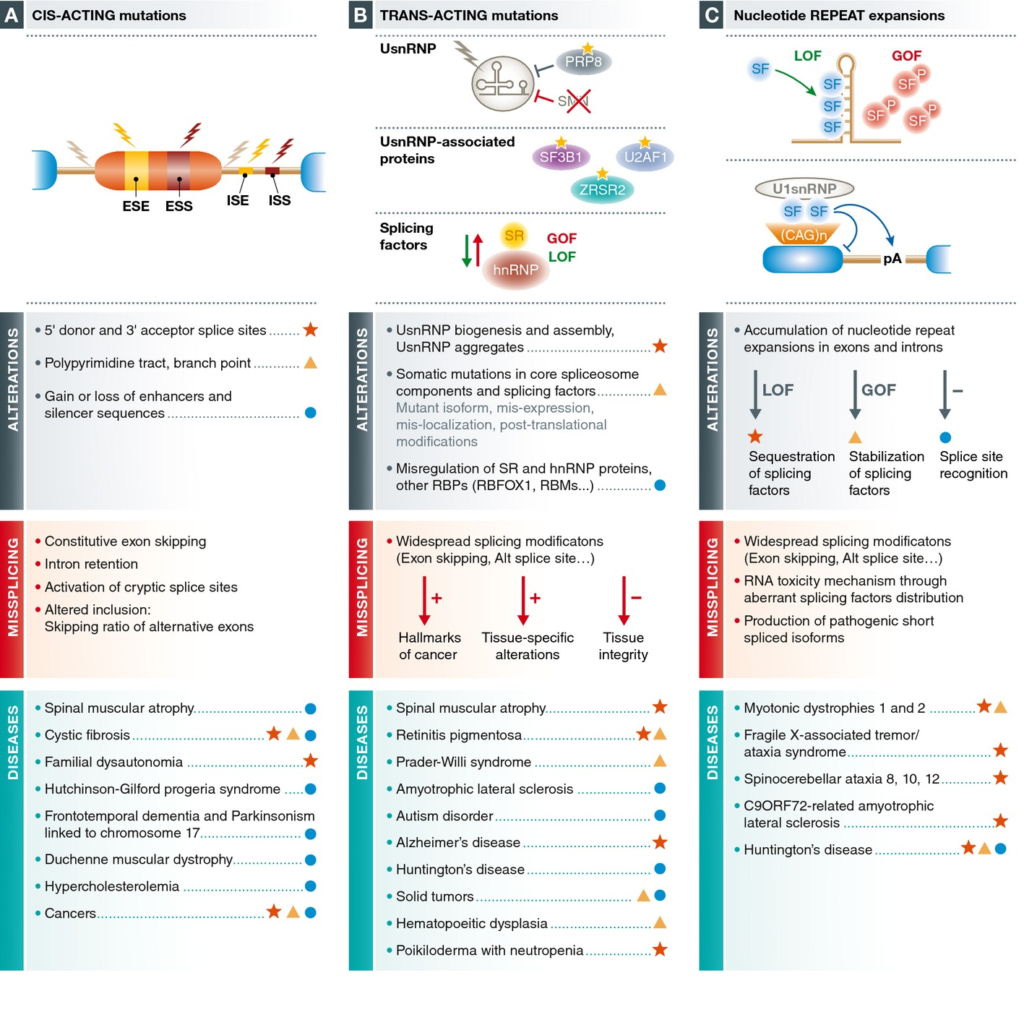
- Mutaciones de un gen que afectan a sus sitios de splicing.
- Cis- regulatory element splicing mutations.
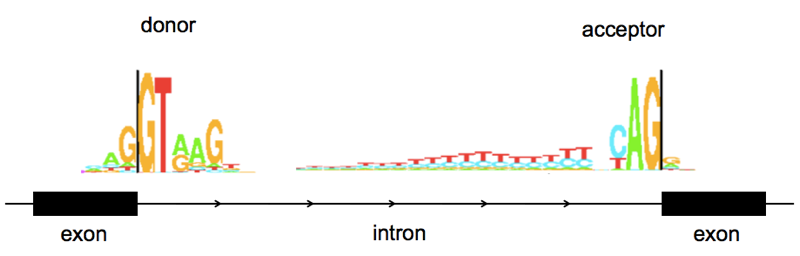
- Mutaciones tipo I. Sitio donador canónico (GT). Suelen anotarse con -1/-2/-3 tras la posición del nucleótido que finaliza el exón.
- Mutaciones tipo II. Variantes intrónicas profundas que producen inclusión exónica críptica.
- Mutaciones tipo III y V. Variantes exónicas que producen alteración del splicing (distintas del sitio aceptor y donador canónico) por afectación de enhacers y silencers.
- Mutaciones tipo IV. Sitio aceptor canónico (AG). Suelen anotarse con +1/+2/+3 tras la posición del nucleótido que inicia el exón.
- Mutaciones que afectan el “branch point” y el tracto de polipirimidina.
- Trans-regulatory element splicing mutations.
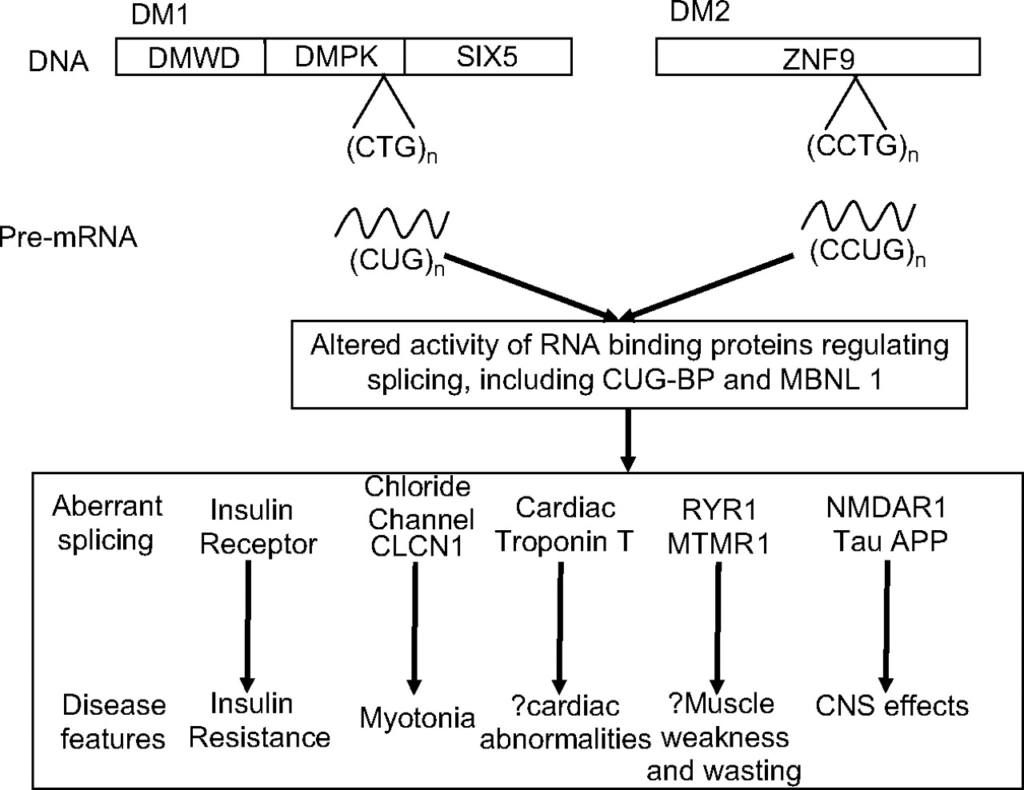
- Mutaciones dinámicas (STR) que dan lugar a productos tóxicos que alteran el normal funcionamiento del splicing y la transcripción.
- Cis- regulatory element splicing mutations.
- Mutaciones que afectan a la maquinaria de regulación del splicing (spliceosoma), dentro de la maquinaria de regulación de la transcripción.
Estrategias para demostrar la patogenicidad.
- Human splicing finder.
- Análisis in-silico.
- Análisis funcional.
332710
{332710:7RSEKJJB},{332710:HEIEWD9Q}
1
vancouver
50
default
3536
https://neuropediatoolkit.org/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22HEIEWD9Q%22%2C%22library%22%3A%7B%22id%22%3A332710%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Daguenet%20et%20al.%22%2C%22parsedDate%22%3A%222015-12%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%201.35%3B%20%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%20style%3D%26quot%3Bclear%3A%20left%3B%20%26quot%3B%26gt%3B%5Cn%20%20%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-left-margin%26quot%3B%20style%3D%26quot%3Bfloat%3A%20left%3B%20padding-right%3A%200.5em%3B%20text-align%3A%20right%3B%20width%3A%201em%3B%26quot%3B%26gt%3B1.%26lt%3B%5C%2Fdiv%26gt%3B%26lt%3Bdiv%20class%3D%26quot%3Bcsl-right-inline%26quot%3B%20style%3D%26quot%3Bmargin%3A%200%20.4em%200%201.5em%3B%26quot%3B%26gt%3BDaguenet%20E%2C%20Dujardin%20G%2C%20Valc%26%23xE1%3Brcel%20J.%20The%20pathogenicity%20of%20splicing%20defects%3A%20mechanistic%20insights%20into%20pre-mRNA%20processing%20inform%20novel%20therapeutic%20approaches.%20EMBO%20reports%20%5BInternet%5D.%202015%20Dec%20%5Bcited%202022%20Nov%203%5D%3B16%2812%29%3A1640%26%23x2013%3B55.%20Available%20from%3A%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.15252%5C%2Fembr.201541116%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.15252%5C%2Fembr.201541116%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%20%20%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20pathogenicity%20of%20splicing%20defects%3A%20mechanistic%20insights%20into%20pre-mRNA%20processing%20inform%20novel%20therapeutic%20approaches%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elisabeth%22%2C%22lastName%22%3A%22Daguenet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gwendal%22%2C%22lastName%22%3A%22Dujardin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juan%22%2C%22lastName%22%3A%22Valc%5Cu00e1rcel%22%7D%5D%2C%22abstractNote%22%3A%22Abstract%20Removal%20of%20introns%20from%20pre-mRNA%20precursors%20%28pre-mRNA%20splicing%29%20is%20a%20necessary%20step%20for%20the%20expression%20of%20most%20genes%20in%20multicellular%20organisms%2C%20and%20alternative%20patterns%20of%20intron%20removal%20diversify%20and%20regulate%20the%20output%20of%20genomic%20information.%20Mutation%20or%20natural%20variation%20in%20pre-mRNA%20sequences%2C%20as%20well%20as%20in%20spliceosomal%20components%20and%20regulatory%20factors%2C%20has%20been%20implicated%20in%20the%20etiology%20and%20progression%20of%20numerous%20pathologies.%20These%20range%20from%20monogenic%20to%20multifactorial%20genetic%20diseases%2C%20including%20metabolic%20syndromes%2C%20muscular%20dystrophies%2C%20neurodegenerative%20and%20cardiovascular%20diseases%2C%20and%20cancer.%20Understanding%20the%20molecular%20mechanisms%20associated%20with%20splicing-related%20pathologies%20can%20provide%20key%20insights%20into%20the%20normal%20function%20and%20physiological%20context%20of%20the%20complex%20splicing%20machinery%20and%20establish%20sound%20basis%20for%20novel%20therapeutic%20approaches.%22%2C%22date%22%3A%222015-12%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.15252%5C%2Fembr.201541116%22%2C%22ISSN%22%3A%221469-221X%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.15252%5C%2Fembr.201541116%22%2C%22collections%22%3A%5B%22AQQ4HSSL%22%5D%2C%22dateModified%22%3A%222025-08-18T18%3A33%3A59Z%22%7D%7D%2C%7B%22key%22%3A%227RSEKJJB%22%2C%22library%22%3A%7B%22id%22%3A332710%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Anna%20and%20Monika%22%2C%22parsedDate%22%3A%222018%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%201.35%3B%20%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%20style%3D%26quot%3Bclear%3A%20left%3B%20%26quot%3B%26gt%3B%5Cn%20%20%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-left-margin%26quot%3B%20style%3D%26quot%3Bfloat%3A%20left%3B%20padding-right%3A%200.5em%3B%20text-align%3A%20right%3B%20width%3A%201em%3B%26quot%3B%26gt%3B1.%26lt%3B%5C%2Fdiv%26gt%3B%26lt%3Bdiv%20class%3D%26quot%3Bcsl-right-inline%26quot%3B%20style%3D%26quot%3Bmargin%3A%200%20.4em%200%201.5em%3B%26quot%3B%26gt%3BAnna%20A%2C%20Monika%20G.%20Splicing%20mutations%20in%20human%20genetic%20disorders%3A%20examples%2C%20detection%2C%20and%20confirmation.%20J%20Appl%20Genetics%20%5BInternet%5D.%202018%20%5Bcited%202021%20July%2012%5D%3B59%283%29%3A253%26%23x2013%3B68.%20Available%20from%3A%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttp%3A%5C%2F%5C%2Flink.springer.com%5C%2F10.1007%5C%2Fs13353-018-0444-7%26%23039%3B%26gt%3Bhttp%3A%5C%2F%5C%2Flink.springer.com%5C%2F10.1007%5C%2Fs13353-018-0444-7%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%20%20%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Splicing%20mutations%20in%20human%20genetic%20disorders%3A%20examples%2C%20detection%2C%20and%20confirmation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Abramowicz%22%2C%22lastName%22%3A%22Anna%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gos%22%2C%22lastName%22%3A%22Monika%22%7D%5D%2C%22abstractNote%22%3A%22Precise%20pre-mRNA%20splicing%2C%20essential%20for%20appropriate%20protein%20translation%2C%20depends%20on%20the%20presence%20of%20consensus%20Bcis%5E%20sequences%20that%20define%20exon-intron%20boundaries%20and%20regulatory%20sequences%20recognized%20by%20splicing%20machinery.%20Point%20mutations%20at%20these%20consensus%20sequences%20can%20cause%20improper%20exon%20and%20intron%20recognition%20and%20may%20result%20in%20the%20formation%20of%20an%20aberrant%20transcript%20of%20the%20mutated%20gene.%20The%20splicing%20mutation%20may%20occur%20in%20both%20introns%20and%20exons%20and%20disrupt%20existing%20splice%20sites%20or%20splicing%20regulatory%20sequences%20%28intronic%20and%20exonic%20splicing%20silencers%20and%20enhancers%29%2C%20create%20new%20ones%2C%20or%20activate%20the%20cryptic%20ones.%20Usually%20such%20mutations%20result%20in%20errors%20during%20the%20splicing%20process%20and%20may%20lead%20to%20improper%20intron%20removal%20and%20thus%20cause%20alterations%20of%20the%20open%20reading%20frame.%20Recent%20research%20has%20underlined%20the%20abundance%20and%20importance%20of%20splicing%20mutations%20in%20the%20etiology%20of%20inherited%20diseases.%20The%20application%20of%20modern%20techniques%20allowed%20to%20identify%20synonymous%20and%20nonsynonymous%20variants%20as%20well%20as%20deep%20intronic%20mutations%20that%20affected%20pre-mRNA%20splicing.%20The%20bioinformatic%20algorithms%20can%20be%20applied%20as%20a%20tool%20to%20assess%20the%20possible%20effect%20of%20the%20identified%20changes.%20However%2C%20it%20should%20be%20underlined%20that%20the%20results%20of%20such%20tests%20are%20only%20predictive%2C%20and%20the%20exact%20effect%20of%20the%20specific%20mutation%20should%20be%20verified%20in%20functional%20studies.%20This%20article%20summarizes%20the%20current%20knowledge%20about%20the%20Bsplicing%20mutations%5E%20and%20methods%20that%20help%20to%20identify%20such%20changes%20in%20clinical%20diagnosis.%22%2C%22date%22%3A%2208%5C%2F2018%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1007%5C%2Fs13353-018-0444-7%22%2C%22ISSN%22%3A%221234-1983%2C%202190-3883%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Flink.springer.com%5C%2F10.1007%5C%2Fs13353-018-0444-7%22%2C%22collections%22%3A%5B%22B8DDX5EW%22%2C%22DK68YYBL%22%5D%2C%22dateModified%22%3A%222025-08-18T18%3A33%3A59Z%22%7D%7D%5D%7D
1.
Daguenet E, Dujardin G, Valcárcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO reports [Internet]. 2015 Dec [cited 2022 Nov 3];16(12):1640–55. Available from: https://www.embopress.org/doi/full/10.15252/embr.201541116
1.
Anna A, Monika G. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J Appl Genetics [Internet]. 2018 [cited 2021 July 12];59(3):253–68. Available from: http://link.springer.com/10.1007/s13353-018-0444-7